发布者:Primerbank 时间:2017-10-09 浏览量:1744
First published: 11 November 2010
DOI: 10.1111/j.1469-0691.2010.03344.x
Cited by (CrossRef): 10 articles
Corresponding author L. Cnops, Department of Clinical Sciences, Kronenburgstraat 43/3, B-2000 Antwerp, Belgium
E-mail:lcnops@itg.be
Clin Microbiol Infect 2011; 17: 1101–1107
Although microscopy remains the reference standard for malaria diagnosis, molecular tools are attracting increasing interest. To improve the detection of mixed infections, we developed a four-primer real-time PCR with four Plasmodiumspecies-specific forward primers, based on the pan-primer design with universal Plasmodium primers as described previously. After validation for analytical sensitivity, specificity and reproducibility, the four-primer PCR was evaluated on 351 blood samples from patients presenting at the outpatient clinic of the Institute of Tropical Medicine (Belgium). With the four-primer PCR, we identified 188 Plasmodium falciparum (Pf), 54Plasmodium vivax (Pv), 52 Plasmodium ovale (Po) and 13Plasmodium malariae (Pm) single infections, 27 mixed infections (14 Pf + Pm; 12 Pf + Po; one Pv + Pm) and 17 negative specimens. We found lower cycle threshold values than with the pan-primer PCR, with a mean difference of 2.23, a higher analytical sensitivity (in asexual parasites/μL: Pf/Pv, 0.02; Po, 0.004; Pm, 0.006) and 15 extra mixed infections. As compared with microscopy, 17 extra mixed infections were detected andPlasmodium species were identified in four microscopy-positive samples in which species identification was not possible. Additionally, the PCR corrected 13 species mismatches between Po and Pv, and in 11 cases detected Pf as a second species that was not identified by microscopy and in five of them was not detected by rapid diagnostic tests (RDTs). PCR confirmed the presence of Pf in 30/46 histidine-rich protein-2-positive samples that were microscopy-negative. We conclude that the presently developed four-primer real-time PCR is complementary to standard malaria diagnostic tests in clinical laboratories, with an added value for simultaneous identification of the four Plasmodium species and the detection of mixed infections.
Malaria is the most important parasitic infection worldwide. Because of the increasing amount of travel to tropical destinations, the number of malaria cases being imported to Europe is rising. Adequate identification of the four Plasmodium species, which differ greatly in their biology and clinical manifestations, is important for successful treatment. In cases of co-infection, diagnosis and subsequent treatment is even more complicated.
Species differentiation is possible by microscopic blood smear examination when conducted by trained and experienced staff. However, low levels of parasitaemia, mixed Plasmodium infections, altered morphology of parasites following (self)treatment and poor smear quality challenge the abilities of even the most skilled microscopists. Some problems can be overcome by the use of rapid diagnostic tests (RDTs), although they have lower sensitivities at low parasite densities and a limited capacity for non-Plasmodium falciparum (Pf) and mixed-species identification. Accordingly, molecular techniques can be applied to resolve discordant results or as reference methods. Many studies have demonstrated increased malaria diagnosis and corrected species identifications by PCR, especially for non-Pf and mixed infections.
Several real-time PCR formats have been successfully applied for malaria diagnosis in non-endemic settings. However, not all formats were designed to differentiate between the fourPlasmodium species, and these appear to be less suitable for the detection of mixed infections than are conventional nested-PCR formats, owing to competitive inhibition. Indeed, universalPlasmodium primers generate competition for PCR reagents in cases of mixed infection, and might give false-negative results for the minor species. To avoid this competition, we developed a new real-time PCR based on the PCR of Rougemont, which we have used before. After validation of the new PCR—hereafter referred to as four-primer PCR—we analysed 351 clinical samples in comparison with the pan-primer PCR (adapted from Rougemont) and with standard microscopy.
DNA was extracted from 200 μL of blood with the Qiagen DNA mini kit (Qiagen Benelux, Venlo, The Netherlands), according to the manufacturer’s guidelines. DNA was eluted with 200 μL of elution buffer (Qiagen).
For the pan-primer PCR, the same universal Plasmodium primer set (plasmo1 and plasmo2) and four species-specific probes (Biolegio, Nijmegen, The Netherlands) (Fig. 1) as previously described were used, except for the Vivprobe, which was made 12 bp shorter and contained a minor groove binder modification. Two duplex reactions were run simultaneously. One 25-μL duplex reaction mix (Pf/Plasmodium vivax (Pv)) contained 5 μL of DNA, Quantitec mix (Qiagen Benelux), 200 nM plasmo1 and plasmo2, 100 nM Falcprobe and 200 nM Vivprobe. In the other duplex reaction mix (Plasmodium malariae (Pm)/Plasmodium ovale (Po)), besides 5 μL of DNA, 1× Quantitec mix, and 500 nM plasmo1 and plasmo2, 320 nM Ovaprobe, 200 nM Malaprobe and 1 mM MgCl2 were added. The PCR program consisted of an initial step of 15 min at 95°C, followed by 50 cycles of 5 s at 95°C, 30 s at 58°C and finally 30 s at 72°C.

Primer and probe sequences. Partial alignment of the Plasmodium falciparum (Pf) (accession number M19172; bp 1067–1209), Plasmodium ovale (Po) (accession number L48987; bp 1079–1223) Plasmodium vivax (Pv) (accession number X13926; bp 1398–1542) and Plasmodium malariae(Pm) (accession number AF488000; bp 1106–1256) 18S rRNA gene around the region of selected primers and probes. The pan-primer PCR uses universal Plasmodium genus forward (FW) (plasmo1) and reverse (plasmo2) primers (bold italic) in combination with four species-specific probes (Falcprobe, Vivprobe, Malaprobe and Ovaprobe) (grey highlights). In the four-primer PCR, four species-specific FW primers (underlined) are used instead of plasmo1, resulting in shorter amplicons (Pf, 100 bp; Pv, 105 bp; Pm, 110 bp; Po, 105 bp).
For the four-primer PCR, newly designed species-specific forward (FW) primers (FWfal, FWviv, FWmal, FWova; Biolegio) were used in combination with the species-specific probes and reverse plasmo2 primer (Fig. 1). Two duplex reactions were run in parallel, each containing 200 nM primers and probes, except for 100 nM FWviv primer and Vivprobe, with 1× Perfecta qPCR Supermix (Quanta Biosciences, Gaithersburg, MD, USA). The PCR was run for 2 min at 95°C, and this was followed by 50 cycles of 15 s at 95°C and 60 s at 60°C. FW primer design was verified with Integrated DNA Technology Oligo Analyzer software (v3.1) (http://eu.idtdna.com/analyzer/Applications/OligoAnalyzer/).
PCRs were run on a SmartCycler II (Cepheid Benelux, Bouwel, Belgium). In every run, the non-template control was negative (cycle threshold (Ct) = 0) and the positive control for each species was positive.
Specificity. Primer specificity was checked in silico by BLAST analysis (http://blast.ncbi.nlm.gov/Blast.cgi) and by gel electrophoresis on PCR products. The analytical specificity of the PCR was tested on six non-Plasmodium blood parasites (Leishmania donovani, Trypanosoma cruzi, Trypanosoma brucei gambiensis, Loa loa, Aspergillus andPneumocystis (based on [14]), and the diagnostic specificity on 30 negative controls derived from individuals suspected of having malaria but with no evidence of Plasmodium infection as demonstrated by microscopy and RDTs.
Analytical sensitivity. The detection limit was determined on serial ten-fold dilutions of high-parasitaemia clinical blood samples of the four Plasmodium species. From each 1/10 blood dilution, DNA was extracted, and the highest dilution with a positive signal indicated the detection limit.
Repeatability and reproducibility. The variation in Ct values was determined for each Plasmodium species in a sample that was processed eight times within the same run (repeatability) or five times in different runs (reproducibility).
The panel of clinical samples collected between 1995 and 2009 consisted of EDTA-anticoagulated blood samples (n = 351) from patients who presented at the outpatient clinic of the Institute of Tropical Medicine (ITM) Antwerp, Belgium, or sent by national laboratories for malaria confirmation at the Central Laboratory of Clinical Biology of the ITM (accredited according to ISO15189:2007). PCR analysis was performed between January 2007 and January 2010, prospectively on fresh samples (stored for a maximum of 1 week at 4°C) for routine diagnosis, or retrospectively on a panel of frozen samples (stored at −70°C). Only the first sample per patient was included in this study. Molecular diagnosis was performed with pan-primer PCR until August 2009, and from September 2009 the new four-primer PCR was applied.
Standard microscopic analysis was carried out on Giemsa-stained smears, and parasite density was expressed as the number of asexual parasites/μL .
For antigen detection, an RDT for qualitative detection of Pf histidine-rich protein-2 (HRP-2) was performed according to the manufacturer’s instructions. Until 2007, the Now Malaria test (Binax, Scarborough, ME, USA), which also detects pan-species aldolase, was used. From 2008, the SD-FK60 Malaria Ag Pf/pan test (Standard Diagnostics, Hagal-Dong, Korea), which detects HRP-2 and pan-species parasite lactate dehydrogenase, was used. All specimens positive by microscopy (n = 305) or RDT alone (n = 46) were examined with both real-time PCRs on the same DNA extract.
Discrepant results between the PCR methods or between microscopy and PCR were assessed with an additional RDT for Pv (SD-FK70; Standard Diagnostics) targeting the Pv-specific parasite lactate dehydrogenase, by external real-time PCR analysis for species confirmation at the laboratory of Leiden (LUMC, Leiden, The Netherlands) , or by supplementary information obtained from species identification in a treatment follow-up sample.
Improved DNA detection was determined by subtracting Ct values measured by the two PCRs, and is indicated by ΔCt. Theoretically, the amplicon amount doubles every cycle (i.e. increases by 1 log2), and one ΔCt therefore corresponds to a two-fold increase in analytical sensitivity. Statistical differences between logarithmic Ct values of the PCRs were determined by paired t-test analysis. The yield in mixed infections was statistically analysed with the two-tailed Fisher exact test.
The newly developed FW primers were specific for each of the fourPlasmodium species (Fig. 1) and were located more closely to the probe hybridization positions, resulting in shorter amplicon lengths. Integrated DNA Technology Oligo analysis approved no self-dimerization or heterodimerization with the reverse primers and probes. BLAST analysis indicated 100% query coverage and maximum identity with the correlated species. Gel electrophoresis provided a single band of the expected length, and no signal in the non-template control.
After optimization of the reaction products and programme, the PCR was validated. No amplification was seen in six non-Plasmodiumcontrols or 30 negative control samples. Analytical sensitivity tests on serial dilutions demonstrated detection limits of 0.02, 0.02, 0.004 and 0.006 parasites/μL for Pf, Pv, Po and Pm, respectively, which corresponds to approximately one to four asexual parasites per unit of blood volume (200 μL). Repeatability and reproducibility testing revealed coefficients of variation lower than 0.8% and 1.3%, respectively, for all Plasmodium species.
Table 1 shows malaria diagnosis for 351 whole blood samples by standard microscopy, pan-primer PCR and four-primer PCR. Overall, more malaria cases were diagnosed with PCR than with microscopy. Among 351 samples, 305 (86.9%) were positive by microscopy, and 331 (94.3%) and 334 (95.2%) with the pan-primer PCR and four-primer PCR, respectively. The numbers of mixed infections detected by microscopy, pan-primer PCR and four-primer PCR were 10, 12 and 27, respectively. The significant increase in yield of mixed infections with the four-primer PCR as compared with the pan-primer PCR (p <0.05) and microscopy (p ≤0.01) involved both Pf/Pm and Pf/Po (14 and 12, respectively). In four samples, identification was possible with both PCRs but not by microscopy (Table 1).
Table 1. Species distribution of Plasmodium spp. according to microscopy, pan-primer PCR and four-primer real-time PCR
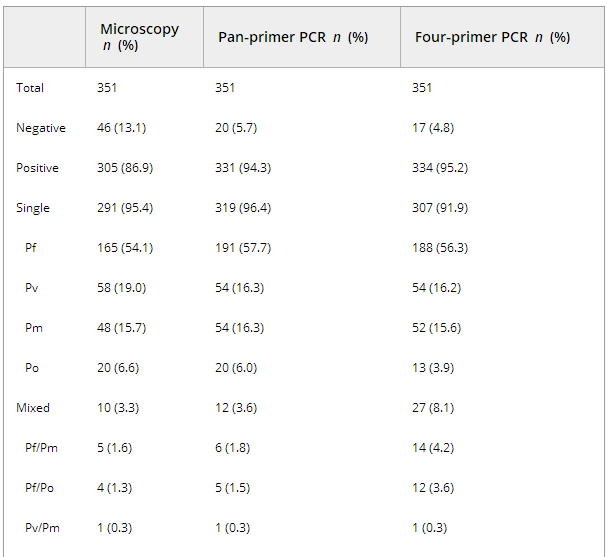
Table 2. Cycle threshold (Ct) values of the pan-primer PCR and four-primer real-time PCR for the four Plasmodium species

Table 3 shows concordant results between both PCR methods for 183 Pf, 54 Pv, 52 Po and 13 Pm single infections, 12 mixed infections and 15 negative samples. In contrast, for 15 samples, DNA amplification of two species was seen with the four-primer PCR but not with the pan-primer PCR (Table 3, footnotes b–e). In those cases, Pf was additionally recognized in seven co-infections with Pm and in two co-infections with Po. All mixed infections could be confirmed by additional tests, except for three patients.
Table 3. Cross-tabulation of species identifications generated by both real-time PCR methods
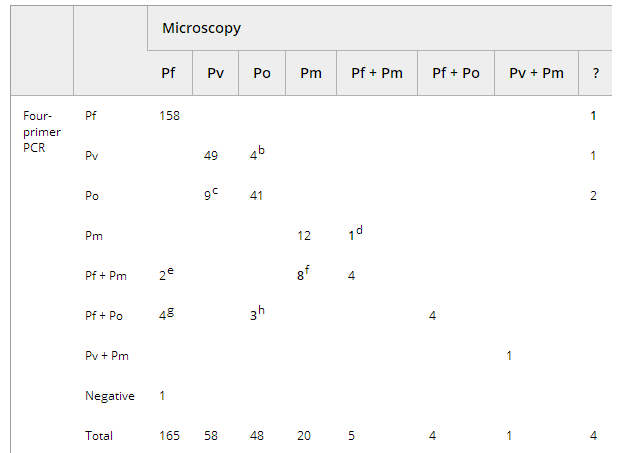
Five samples that were Pf-positive with the four-primer PCR (Ct values >38.8), were missed with the pan-primer PCR (Table 3, footnote a). All five samples were HRP-2-positive and in one sample six parasites/μL were seen with microscopy. For two samples, only the pan-primer PCR (Ct values >40.5) and RDT were Pf-positive, microscopy and the four-primer PCR giving negative results (Table 3, footnote f).
Comparison of the four-primer PCR and microscopy
Table 4 shows the four-primer PCR and microscopy results. Single species identifications showed concordant results for 158 Pf, 49 Pv, 41 Po and 12 Pm samples. In nine microscopically detected mixed infections, DNA of the same two species was co-amplified (four Pf + Pm; four Pf + Po; one Pv + Pm). Sixteen HRP-2-positive samples were negative by both PCR and microscopy (Table 4, footnote j).
Table 4. Microscopic vs. four-primer real-time PCR analysis
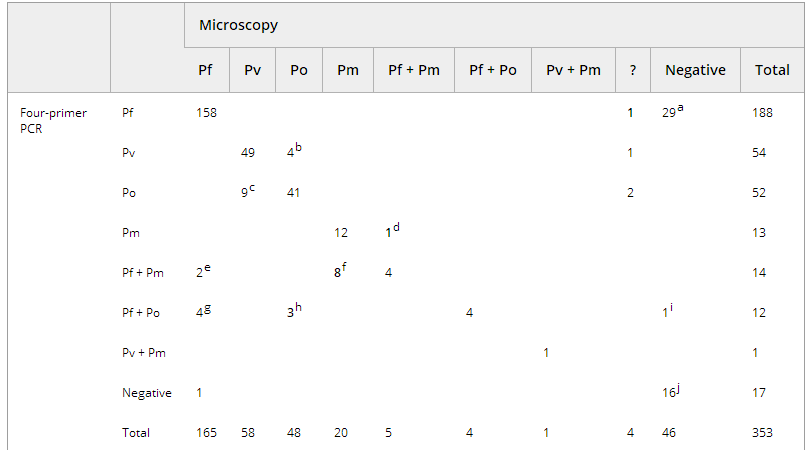

Species identification by microscopy was not possible in four cases, although parasites were seen. PCR analysis revealed Pf (n = 1), Pv (n = 1) and Po (n = 2) in those samples.
Species mismatches were seen for 13/103 (12.6%) Po or Pv infections (Table 4, footnotes b and c). Examination with an additional Pv-specific RDT or external PCR analysis confirmed the identifications in favour of the PCR.
For 17 samples (Table 4, footnotes e–h), microscopic examination resulted in single species identifications, and PCR detected two species as confirmed by additional tests on eight samples in favour of the PCR. Pf was missed by microscopy in 11 samples, and it was co-amplified (eight Pf + Pm; three Pf + Po) as a minor species with Ct values >33. The presence of Pf was confirmed by RDT in 6/11 samples.
In one Pf/Pm mixed infection diagnosed by microscopy, Pf was not detected by four-primer PCR and external PCR or RDT analysis. For another sample, no DNA amplification was seen, and microscopy demonstrated 13 parasites/μL for Pf. Finally, 30 of the 46 microscopically negative but HRP-2-positive samples (Table 3, footnotes h and i) gave a positive PCR signal, with a mean Ct value of 36.3 (range: 27.3–42.4).
This study demonstrates that the four-primer real-time PCR is a powerful tool for malaria diagnosis. It performed excellently with regard to specificity, sensitivity and reproducibility, and, unlike in other studies , was validated on a large number of positive clinical samples in a non-endemic setting. Our real-time format simultaneously detects four Plasmodium species with a turn-around time of <3 h, which is faster than the simplex or nested-PCR methods used by others .
We showed that the four-primer PCR performed better than the pan-primer PCR. First, we consistently obtained lower Ct values, resulting in a more than four-fold increased analytical sensitivity. Second, the detection limit for all species was lower than described for the pan-primer PCR (0.2–2 parasites/μL) and other real-time PCR methods . Third, more mixed infections were detected. The improved amplification is mainly attributable to the limited competition for primers, demonstrating that accurate primer and probe design is essential during PCR development.
Many studies have reported that the actual malaria prevalence estimated by PCR is higher than previously determined by microscopy . A report based on 13 studies that compared microscopy with PCR described 11% more imported malaria cases, and we obtained 8.3% more malaria diagnoses.
PCR is clearly of value for species identification in cases where microscopy fails, as demonstrated in this article. First, PCR corrected 11.4% of Po/Pv cases misdiagnosed by microscopy, a typical species mismatch that has also been seen by others . Second, species identification was possible by PCR but not by microscopy in 1.3% of the microscopy-positive samples. One identification revealed Pf, the most hazardous species. Comparable conclusions were drawn in a report based on ten studies comparing thin slide examinations and PCR, in which 17.2% microscopically determined identifications were corrected by PCR, mainly for non-Pf malaria, and less so for Pf malaria .
It is of note that in one microscopy-positive sample, no PCR amplification was seen, probably because of DNA degradation or inhibition of the PCR amplification. The mainly retrospective nature of our study did not allow exploration of all discordant results.
The most remarkable result from this study is the capacity of the four-primer PCR to detect mixed infections. Generally, the proportion of mixed infections detected by molecular methods in non-endemic settings is higher than by microscopy, varying from 5% up to 12% . In this study, a two-fold higher prevalence of mixed infections was found as compared with microscopy, and even as compared with the pan-primer PCR. This is of concern particularly in cases of missed Pf co-infections, which have substantial implications for the treatment and clinical course of the disease . In this study, Pf was identified by the four-primer PCR as the second species in 11 mixed infections. Five of these were missed by RDT analysis and microscopy, probably because of the low parasite density.
We have described PCR analysis of specimens that were picked up by microscopy or by RDT alone. The limitation of this approach is that no negative samples were analysed, so we cannot calculate the increase in sensitivity by analysing all samples of patients suspected of having malaria with PCR without regarding microscopy or RDT results, although we have anecdotal evidence for it.
Analysis of samples that were positive only by RDT illustrates PCRs capacity to demonstrate the presence of Pf parasites in (partly) treated patients. In about two-thirds of samples that were only HRP-2-positive, PCR confirmed Pf and thus excluded the false-positive reactions that can be obtained with RDTs .
PCR is also of value for species identification in (partly) treated patients with non-Pf or mixed infections that are detected but not identified by RDTs. The role for PCR in treatment follow-up and prediction of treatment failure needs further research. Altogether, we conclude that the four-primer real-time PCR allows identification of the four Plasmodium species in single and mixed infections, and is of added value in reference settings.